Getting started
tmap is a very fast visualization library for large, high-dimensional data sets. Currently, tmap is available for Python.
NEW: We now provide a web-service that allows for the creaton of TMAP visualizations for small chemical data sets.
Supported Environments
| Language | Operating System | Status |
|---|---|---|
| Python | Linux | Available |
| Windows | Available1 | |
| macOS | Available | |
| R | Available2 |
1Works with
WSL
2Through the reticulate R interface to
Python.
Installation
tmap is installed using the conda package manager. Don't have conda? Download miniconda.
conda install -c tmap tmapWe suggest using faerun to plot the data layed out by tmap. But you can of course also use matplotlib (which might be to slow for large data sets and doesn't provide interactive features). Following, an example plot produced with tmap and matplotlib.
pip install faerun
# pip install matplotlibLaying out a Simple Graph
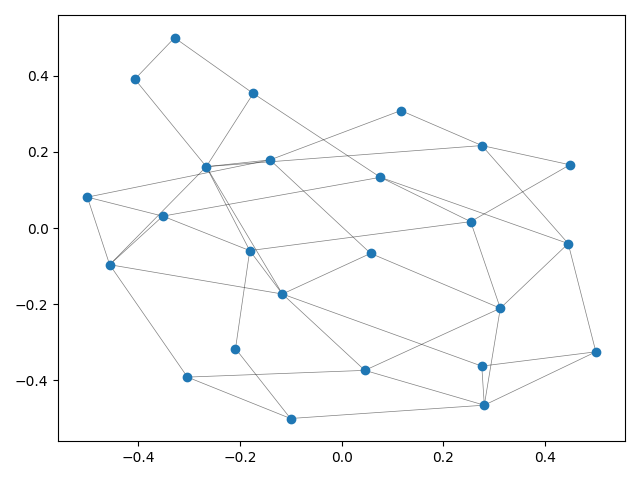
Even though TMAP is mainly targeted at tasks consisting of laying out very large data sets as trees, the simplest usage example is laying out a graph.
This example produces a matplotlib plot displaying the graph.
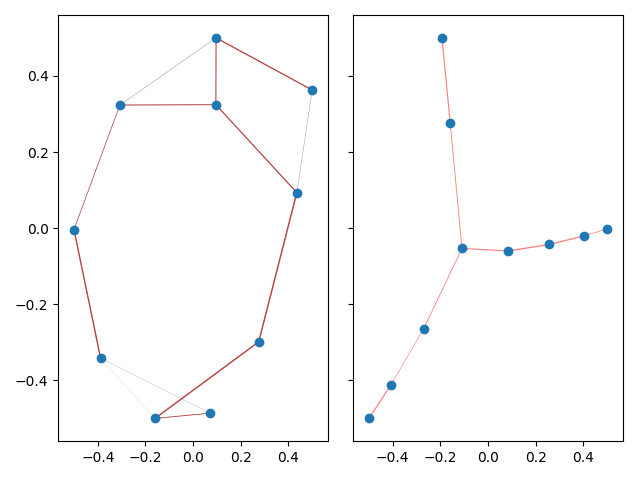
When laying out large graphs, it might be useful to discard some
edges in order to create a more interpretable and visually
pleasing layout. This is achieved using the (default) argument
create_mst=True. Following,
this is exemplified on a small graph.
This example produces a matplotlib plot displaying the graph and it's spanning tree. In the plot on the left, the spanning tree is highlighted by coloring the edges red. The thickness of the edges is inversely related to the edge weight.
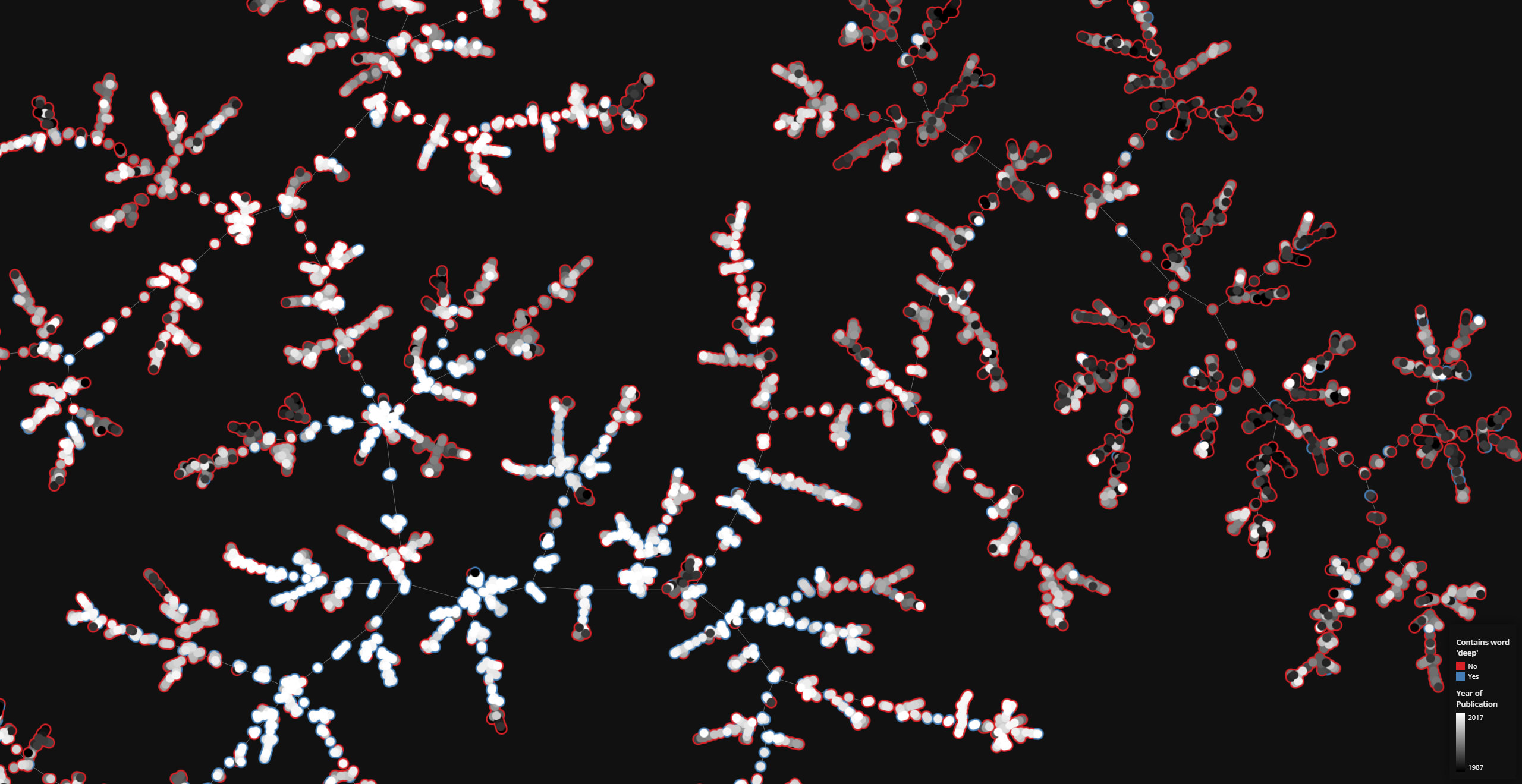
On a highly connected graph with 1000 vertices, the advantages of the tree visualizaton method applied by TMAP become obvious.
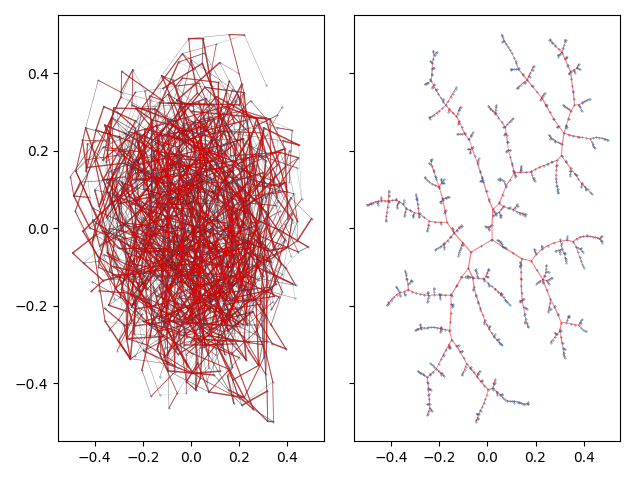
There are a wide array of options to tune the final tree layout to your linking. See Layout for the descriptions of all available parameters.
MinHash
To enable the visualization of larger data sets, it is necessary to speed up the k-nearest neighbor graph generation. While in general, any approach can be used to create this nearest neighbor graph (see Laying out a Simple Graph), tmap provides a built-in LSH Forest data structure, which enables extremely fast k-nearest neighbor queries.
In order to index data in the LSH forest data structure, it has to be hashed using a locality sensitive scheme such as MinHash.
tmap includes the two classes
Minhash and
LSHForest for fast k-nearest
neighbor search.
The following example shows how to use the
Minhash class to estimate
Jaccard distances.
python minhash.py
0.390625
0.140625LSH Forest
The hashes generated by
Minhash can be indexed using
LSHForest for fast k-nearest
neighbor retreival.
python lsh_forest.py
Generating the data took 1164.400289999321ms.
Encoding the data took 155.53330996772274ms.
Adding the data took 17.799988971091807ms.
Indexing took 4.7386749647557735ms.
The kNN search took 0.26249000802636147ms.
After indexing the data, the 10 nearest neighbor search on a
million 1,000-dimensional vectors took ~0.32ms. In addition, the
LSHForest class also supports
the parallelized generation of a k-nearest neighbor graph using
the method get_knn_graph().
python lsh_forest_knng.py
Generating the data took 1171.7670540092513ms.
Adding the data took 189.09973296104ms.
Indexing took 5.959620990324765ms.
The kNN search took 311.5222750348039ms.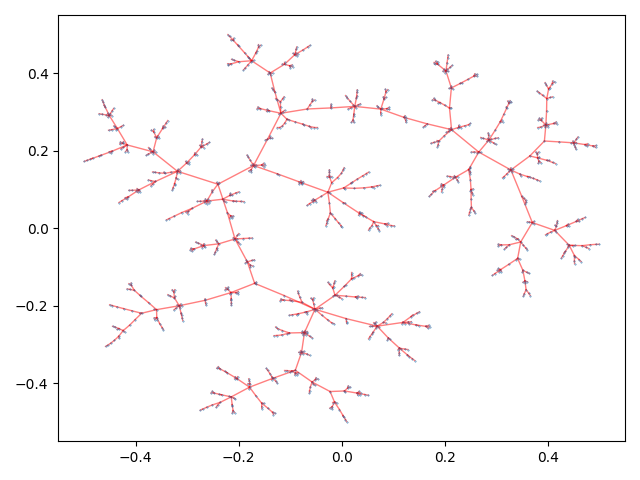
Using matplotlib / pyplot has tow main disadvantages: It is slow and does not yield interactive plots. For this reason, we suggest to use the Python package Faerun for large scale data sets. Faerun supports millions of data points in web-based visualizations.
Together with tmap, Faerun can easily create visualizations of more than 10 million data points including associated web links and structure drawings for high dimensional chemical data sets within an hour.
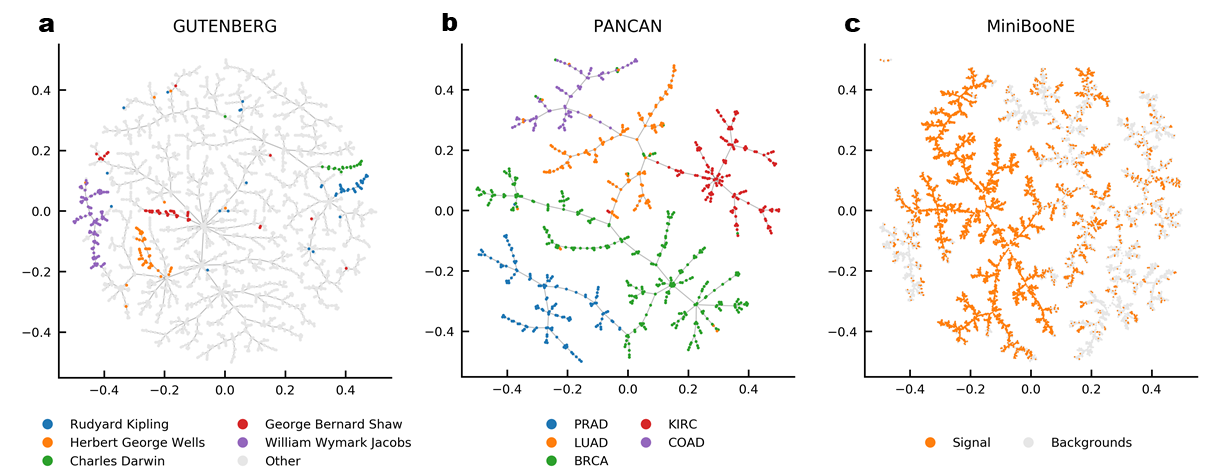
Examples
COIL 20
Data set dimensions: 1,440×16,384

Source
MNIST
Data set dimensions: 70,000×764

Source
Fashion MNIST
Data set dimensions: 70,000×764
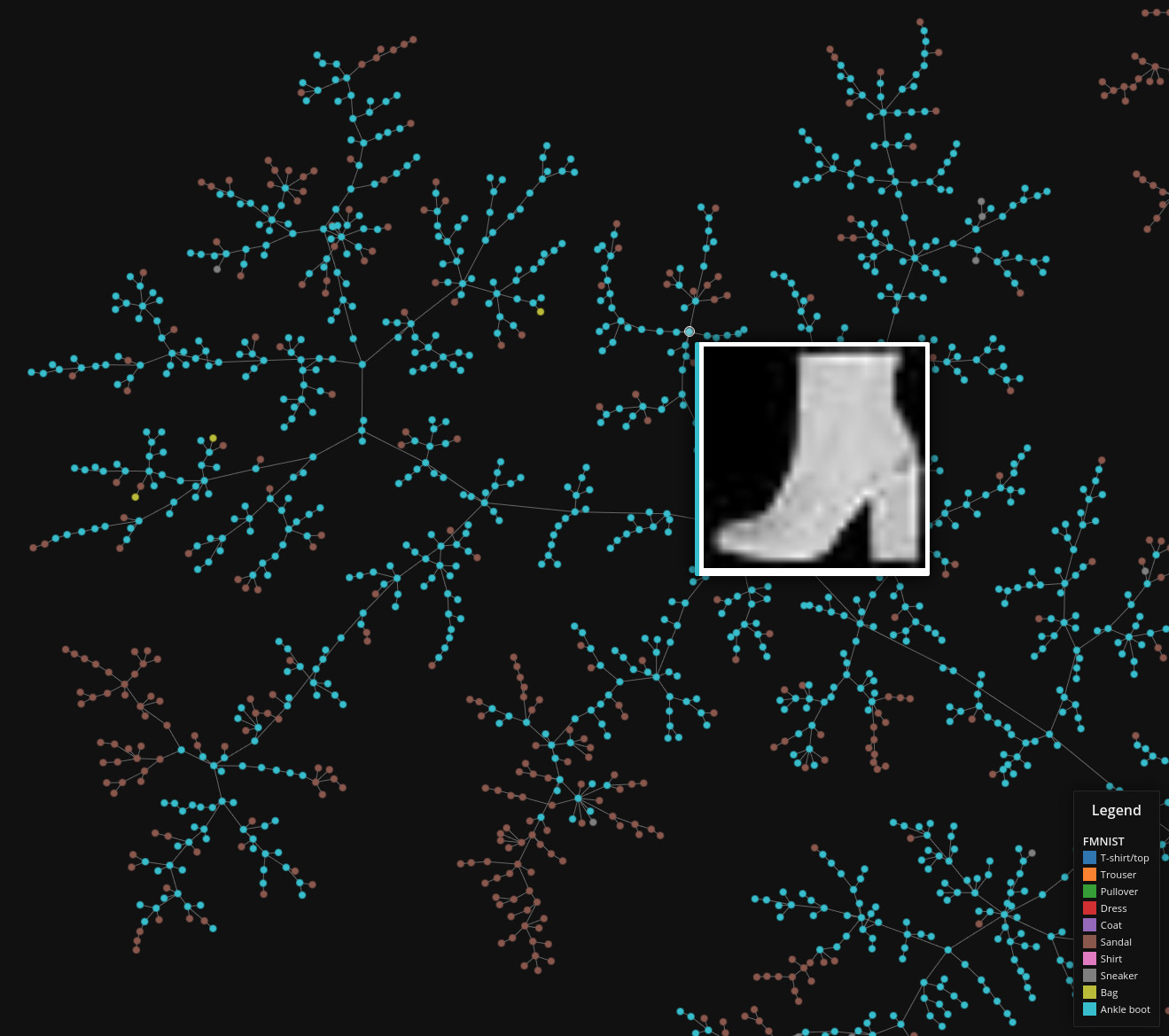
Source
ChEMBL
Data set dimensions: 1,159,881×232
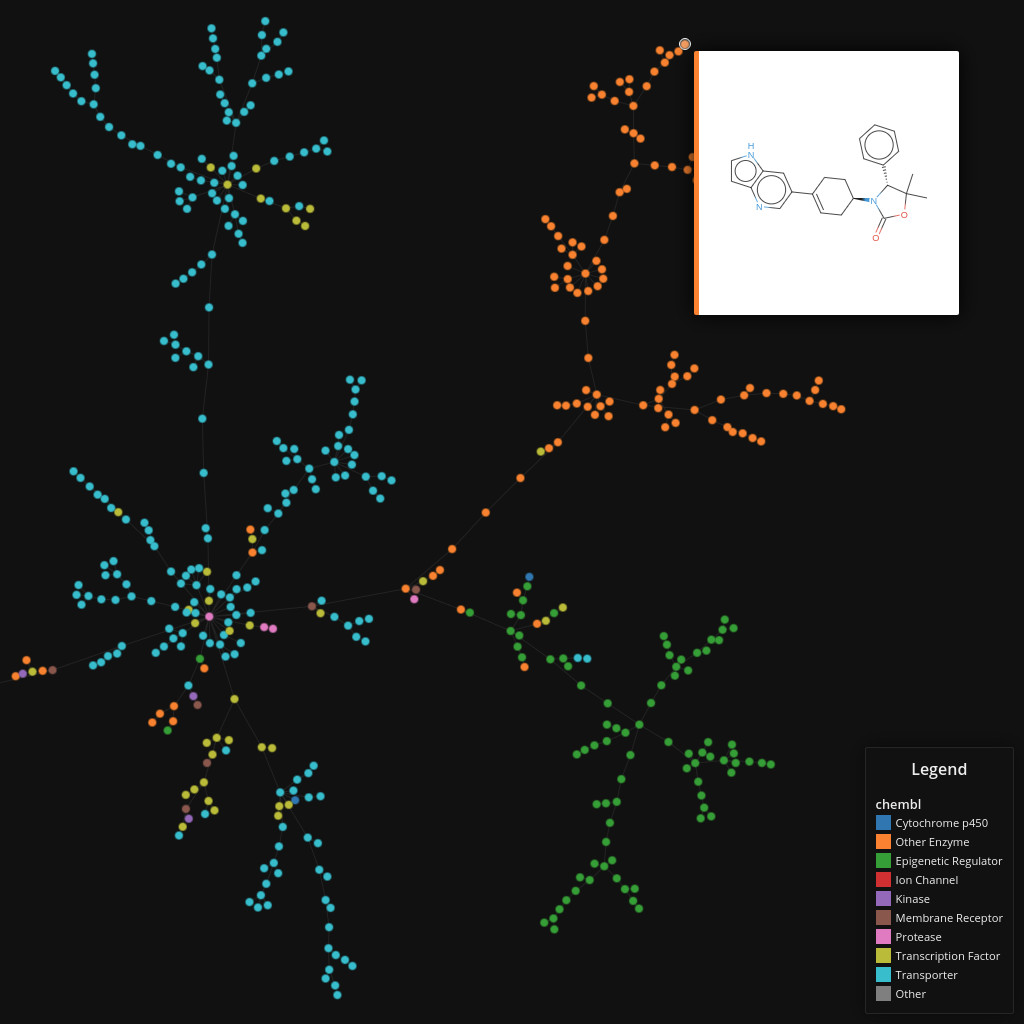
Source
FDB17 and ChEMBL
Data set dimensions: 11,261,085×232
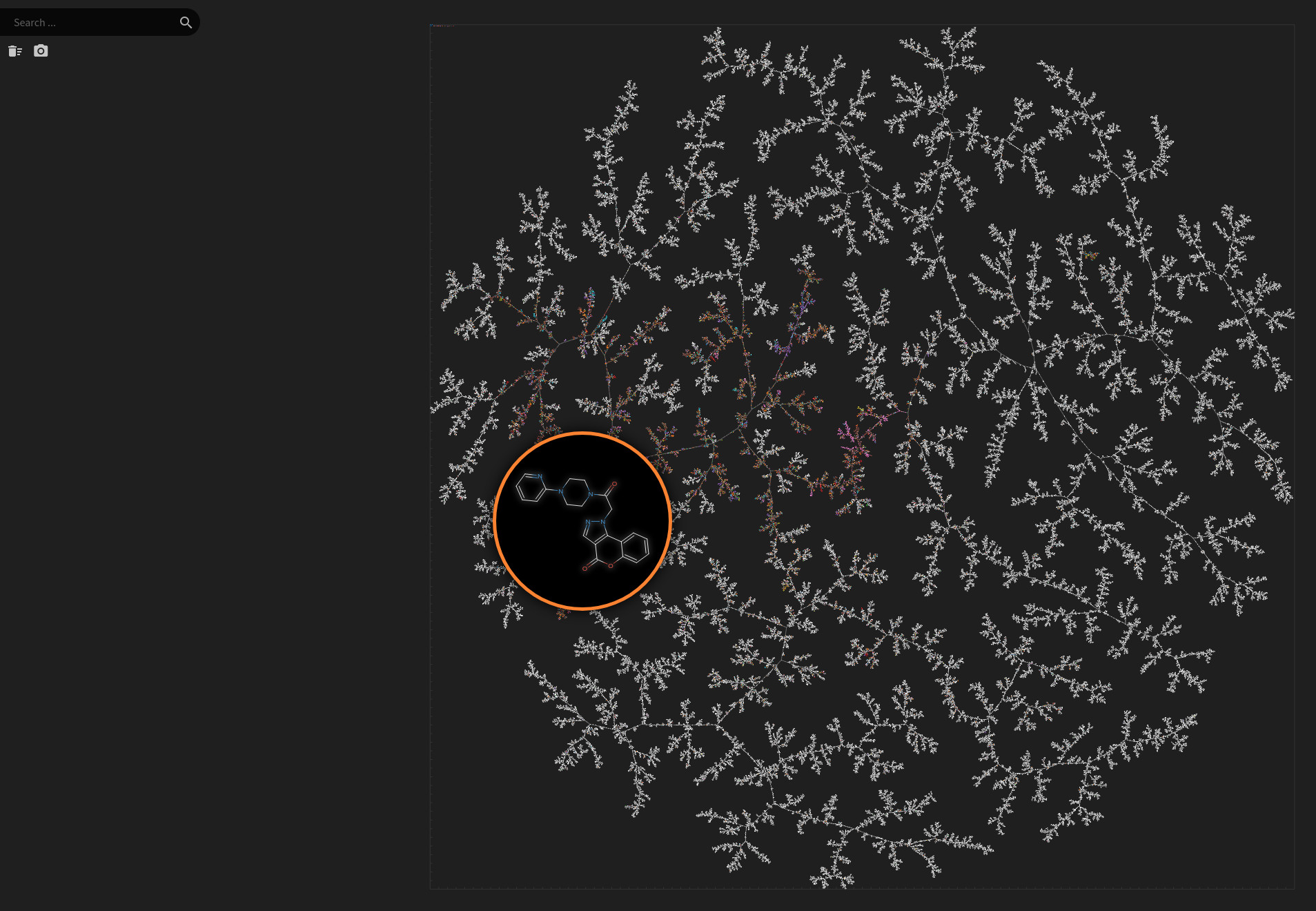
Natural Product Atlas
Data set dimensions: 24,594×232
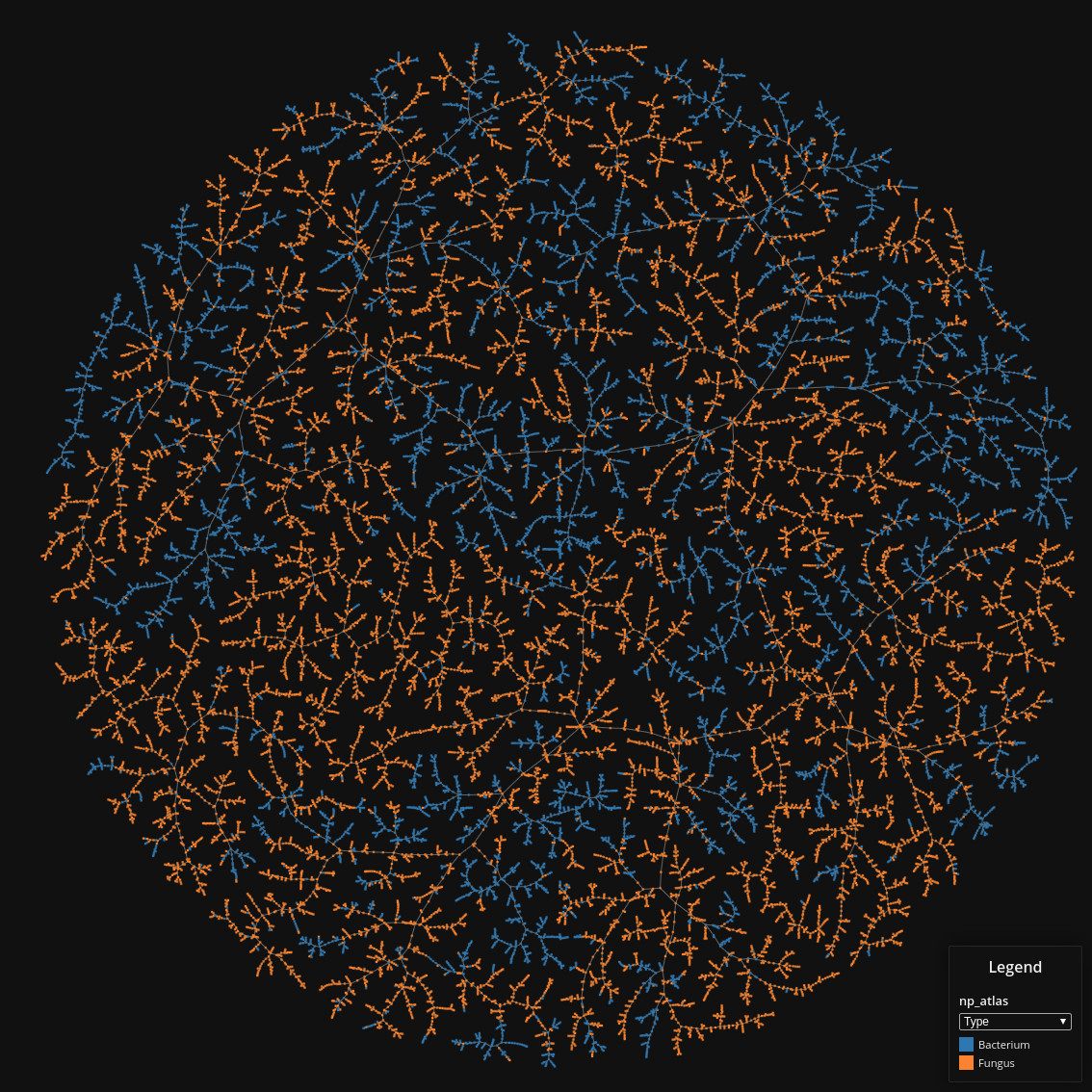
Source
DSSTox
Data set dimensions: 848,816×232
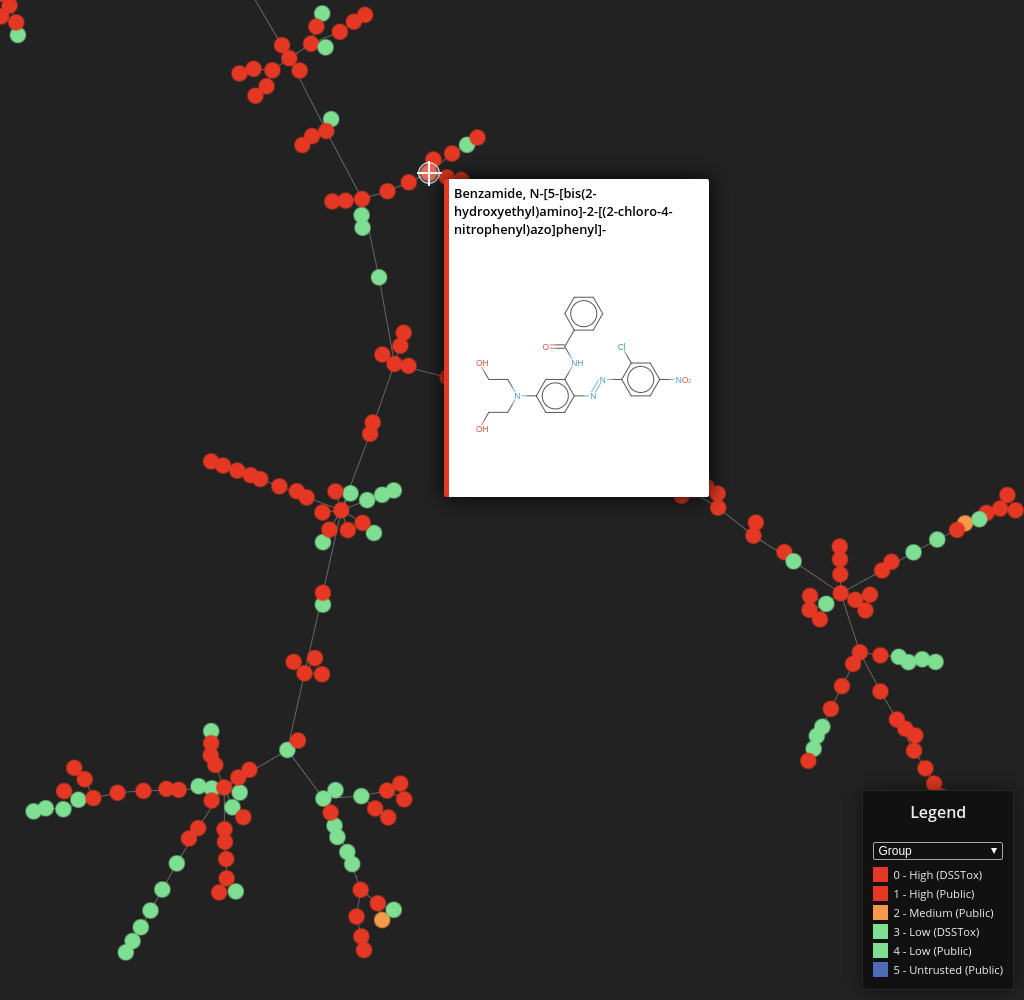
Source
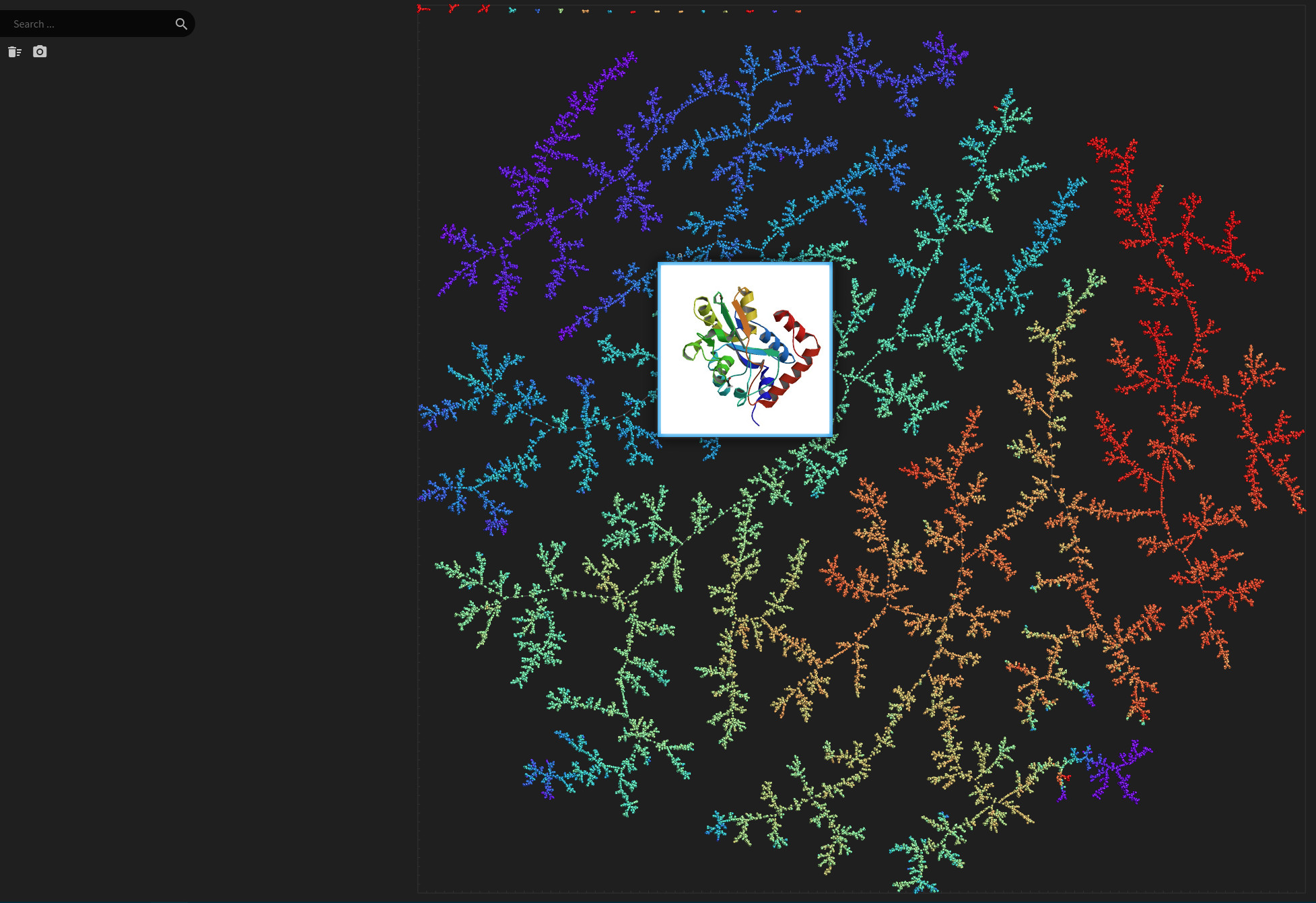
Protein DataBank
Data set dimensions: 131,236×136
RNA Sequencing Data
Data set dimensions: 800×1,022
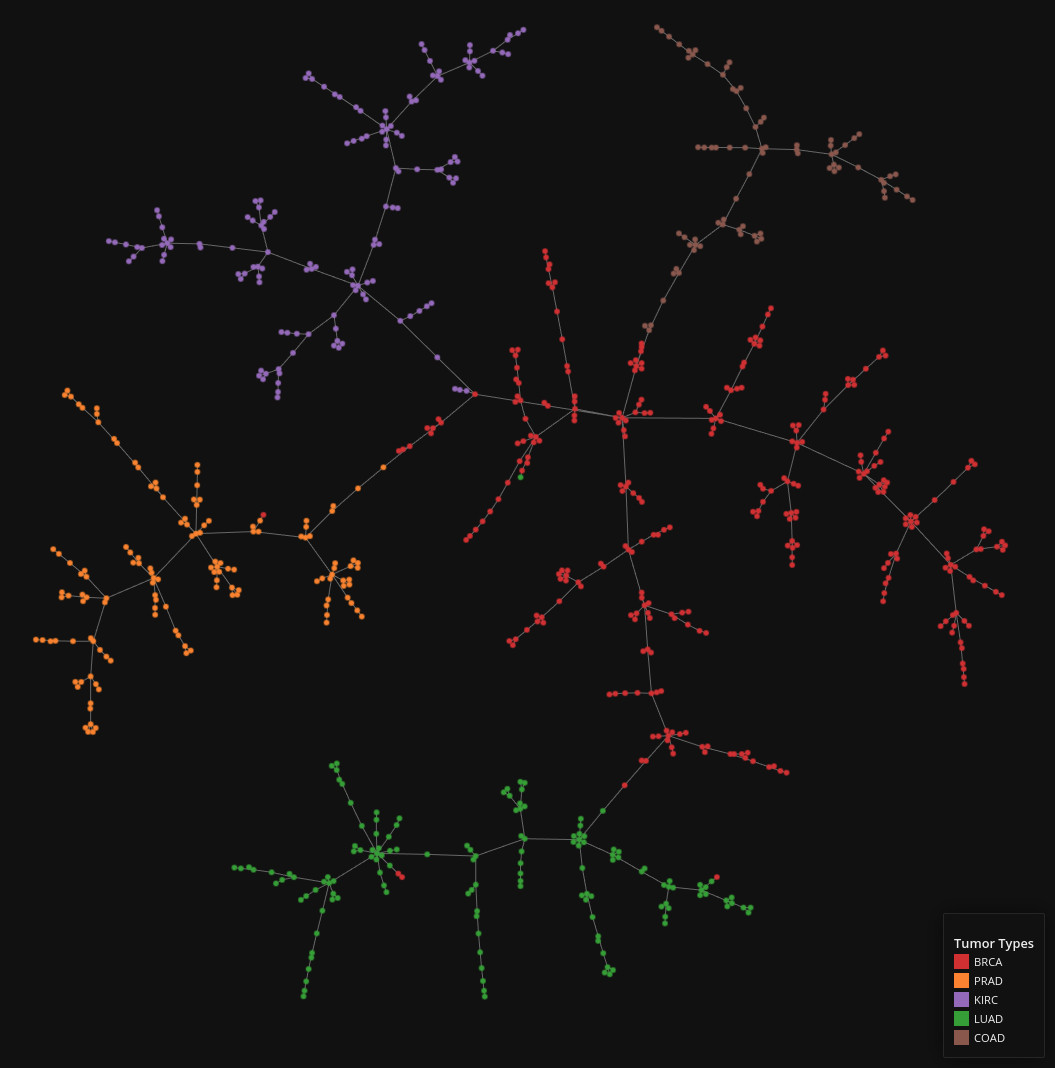
Source
ProteomeHD
Data set dimensions: 5,013×5,013
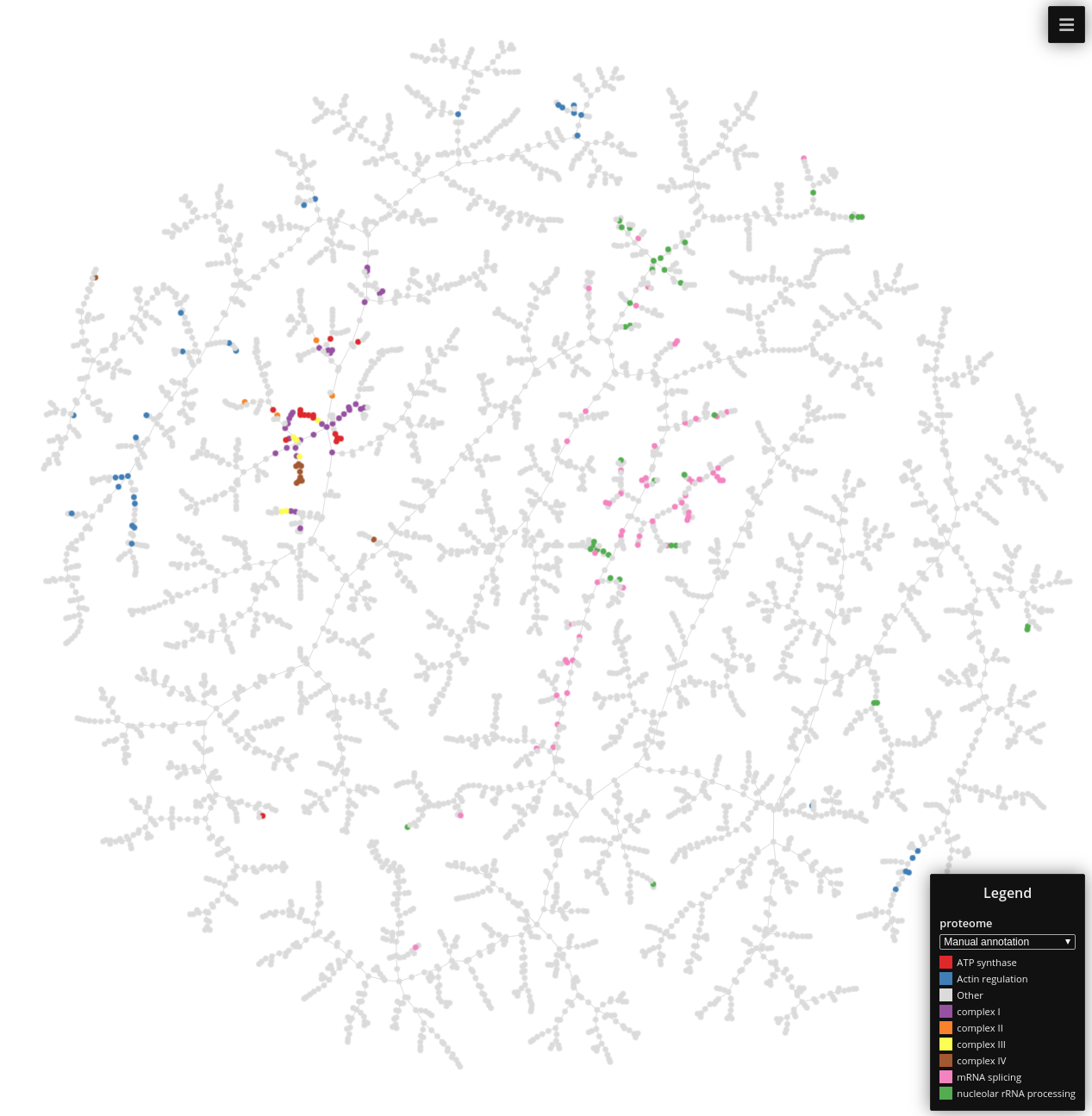
Source
PubMed Central
Data set dimensions: 327,628×1,633,762
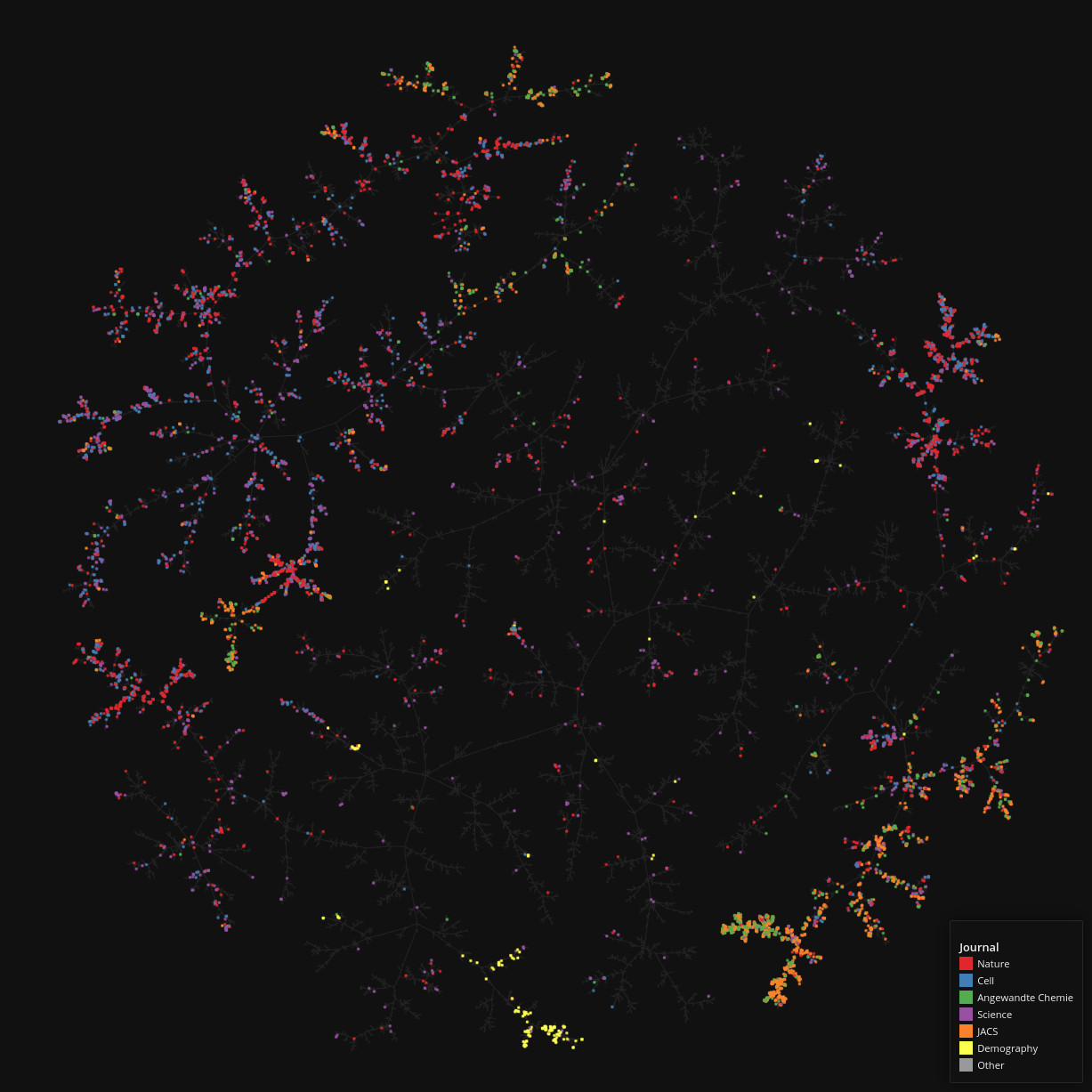
Source
MiniBooNE
Data set dimensions: 130,065×50

Source
Gutenberg
Data set dimensions: 3,036×1,217,078

Source
NIPS Conference Papers
Data set dimensions: 7,241×225,423

Source
Drugbank
Data set dimensions: 9,300×2^32
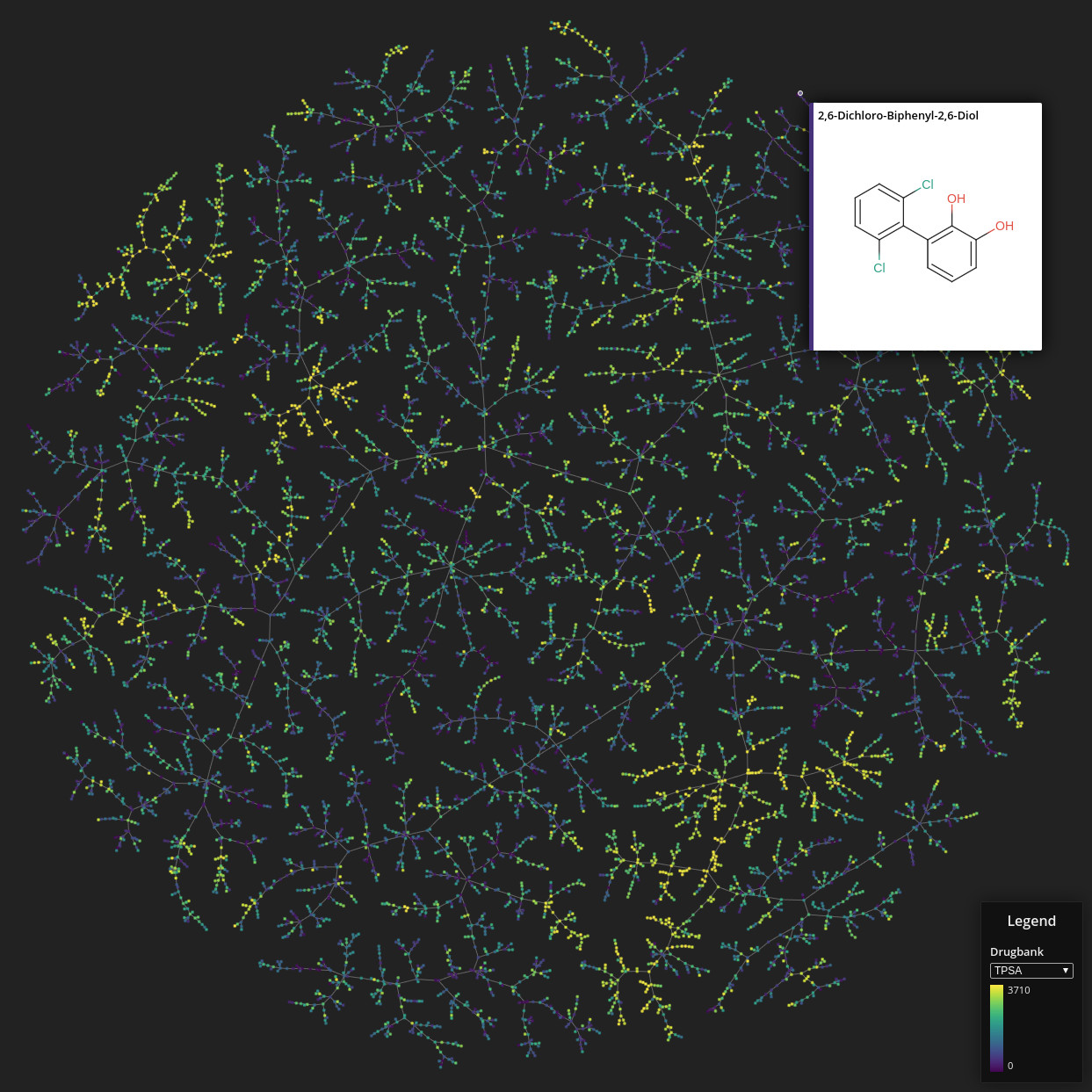
Source
Flowcytometry
Data set dimensions: 436,877×14

Source
moleculenet.ai
Quantum Mechanics
- QM8Source Electronic spectra and excited state energy of small molecules calculated by multiple quantum mechanic methods.
- QM9source Geometric, energetic, electronic and thermodynamic properties of DFT-modelled small molecules.
Physical Chemistry
- ESOLsource Water solubility data(log solubility in mols per litre) for common organic small molecules.
- FreeSolvsource Experimental and calculated hydration free energy of small molecules in water.
- Lipophilicitysource Experimental results of octanol/water distribution coefficient(logD at pH 7.4).
Biophysics
- PCBA (Due to the number of variables in this data set, please run the linked script to generated the visualizations locally) Selected from PubChem BioAssay, consisting of measured biological activities of small molecules generated by high-throughput screening.
- MUVsource Subset of PubChem BioAssay by applying a refined nearest neighbor analysis, designed for validation of virtual screening techniques.
- HIVsource Experimentally measured abilities to inhibit HIV replication.
- PDBbindsource Binding affinities for bio-molecular complexes, both structures of proteins and ligands are provided.
- BACEsource Quantitative (IC50) and qualitative (binary label) binding results for a set of inhibitors of human β-secretase 1(BACE-1).
Physiology
- BBBPsource Binary labels of blood-brain barrier penetration(permeability).
- Tox21source Qualitative toxicity measurements on 12 biological targets, including nuclear receptors and stress response pathways.
- ToxCastsource Toxicology data for a large library of compounds based on in vitro high-throughput screening, including experiments on over 600 tasks.
- SIDERsource Database of marketed drugs and adverse drug reactions (ADR), grouped into 27 system organ classes.
- ClinToxsource Qualitative data of drugs approved by the FDA and those that have failed clinical trials for toxicity reasons.
Documentation
Minhash
-
class
tmap.Minhash(self: tmap.Minhash, d: int=128, seed: int=42, sample_size: int=128) → None¶ -
A generator for MinHash vectors that supports binary, indexed, string and also
intandfloatweighted vectors as input.Constructor for the class
Minhash.- Keyword Arguments
-
-
d (
int) – The number of permutations used for hashing -
seed (
int) – The seed used for the random number generator(s) -
sample_size (
int) – The sample size when generating a weighted MinHash
-
-
batch_from_binary_array(self: tmap.Minhash, arg0: List[tmap.VectorUchar]) → List[tmap.VectorUint]¶ -
Create MinHash vectors from binary arrays (parallelized).
- Parameters
-
vec (
ListofVectorUchar) – A list of vectors containing binary values - Returns
-
A list of MinHash vectors
- Return type
-
ListofVectorUint
-
batch_from_int_weight_array(self: tmap.Minhash, arg0: List[tmap.VectorUint]) → List[tmap.VectorUint]¶ -
Create MinHash vectors from
intarrays, where entries are weights rather than indices of ones (parallelized).- Parameters
-
vec (
ListofVectorUint) – A list of vectors containingintvalues - Returns
-
A list of MinHash vectors
- Return type
-
ListofVectorUint
-
batch_from_sparse_binary_array(self: tmap.Minhash, arg0: List[tmap.VectorUint]) → List[tmap.VectorUint]¶ -
Create MinHash vectors from sparse binary arrays (parallelized).
- Parameters
-
vec (
ListofVectorUint) – A list of vectors containing indices of ones in a binary array - Returns
-
A list of MinHash vectors
- Return type
-
ListofVectorUint
-
batch_from_string_array(self: tmap.Minhash, arg0: List[List[str]]) → List[tmap.VectorUint]¶ -
Create MinHash vectors from string arrays (parallelized).
- Parameters
-
vec (
ListofListofstr) – A list of list of strings - Returns
-
A list of MinHash vectors
- Return type
-
ListofVectorUint
-
batch_from_weight_array(self: tmap.Minhash, vecs: List[tmap.VectorFloat], method: str='ICWS') → List[tmap.VectorUint]¶ -
Create MinHash vectors from
floatarrays (parallelized).- Parameters
-
vec (
ListofVectorFloat) – A list of vectors containingfloatvalues - Keyword Arguments
-
method (
str) – The weighted hashing method to use (ICWS or I2CWS) - Returns
-
A list of MinHash vectors
- Return type
-
ListofVectorUint
-
from_binary_array(self: tmap.Minhash, arg0: tmap.VectorUchar) → tmap.VectorUint¶ -
Create a MinHash vector from a binary array.
- Parameters
-
vec (
VectorUchar) – A vector containing binary values - Returns
-
A MinHash vector
- Return type
-
VectorUint
-
from_sparse_binary_array(self: tmap.Minhash, arg0: tmap.VectorUint) → tmap.VectorUint¶ -
Create a MinHash vector from a sparse binary array.
- Parameters
-
vec (
VectorUint) – A vector containing indices of ones in a binary array - Returns
-
A MinHash vector
- Return type
-
VectorUint
-
from_string_array(self: tmap.Minhash, arg0: List[str]) → tmap.VectorUint¶ -
Create a MinHash vector from a string array.
- Parameters
-
vec (
Listofstr) – A vector containing strings - Returns
-
A MinHash vector
- Return type
-
VectorUint
-
from_weight_array(self: tmap.Minhash, vec: tmap.VectorFloat, method: str='ICWS') → tmap.VectorUint¶ -
Create a MinHash vector from a
floatarray.- Parameters
-
vec (
VectorFloat) – A vector containingfloatvalues - Keyword Arguments
-
method (
str) – The weighted hashing method to use (ICWS or I2CWS) - Returns
-
A MinHash vector
- Return type
-
VectorUint
-
get_distance(self: tmap.Minhash, arg0: tmap.VectorUint, arg1: tmap.VectorUint) → float¶ -
Calculate the Jaccard distance between two MinHash vectors.
- Parameters
-
-
vec_a (
VectorUint) – A MinHash vector -
vec_b (
VectorUint) – A MinHash vector
-
- Returns
-
floatThe Jaccard distance
-
get_weighted_distance(self: tmap.Minhash, arg0: tmap.VectorUint, arg1: tmap.VectorUint) → float¶ -
Calculate the weighted Jaccard distance between two MinHash vectors.
- Parameters
-
-
vec_a (
VectorUint) – A weighted MinHash vector -
vec_b (
VectorUint) – A weighted MinHash vector
-
- Returns
-
floatThe Jaccard distance
LSHForest
-
class
tmap.LSHForest(self: tmap.LSHForest, d: int=128, l: int=8, store: bool=True, file_backed: bool=False, weighted: bool=False) → None¶ -
A LSH forest data structure which incorporates optional linear scan to increase the recovery performance. Most query methods are available in parallelized versions named with a
batch_prefix.Constructor for the class
LSHForest.- Keyword Arguments
-
-
d (
int) – The dimensionality of the MinHashe vectors to be added -
l (
int) – The number of prefix trees used when indexing data -
store (
bool) – Store the data for enhanced querying -
file_backed (
bool) – Whether to store the data on disk rather than in main memory (experimental)
-
-
add(self: tmap.LSHForest, arg0: tmap.VectorUint) → None¶ -
Add a MinHash vector to the LSH forest.
- Parameters
-
vecs (
VectorUint) – A MinHash vector that is to be added to the LSH forest
-
batch_add(self: tmap.LSHForest, arg0: List[tmap.VectorUint]) → None¶ -
Add a list MinHash vectors to the LSH forest (parallelized).
- Parameters
-
vecs (
ListofVectorUint) – A list of MinHash vectors that is to be added to the LSH forest
-
batch_query(self: tmap.LSHForest, arg0: List[tmap.VectorUint], arg1: int) → List[tmap.VectorUint]¶ -
Query the LSH forest for k-nearest neighbors (parallelized).
- Parameters
-
-
vecs (
ListofVectorUint) – The query MinHash vectors -
k (
int) – The number of nearest neighbors to be retrieved
-
- Returns
-
The results of the queries
- Return type
-
ListofVectorUint
-
clear(self: tmap.LSHForest) → None¶ -
Clears all the added data and computed indices from this
LSHForestinstance.
-
get_all_distances(self: tmap.LSHForest, arg0: tmap.VectorUint) → tmap.VectorFloat¶ -
Calculate the Jaccard distances of a MinHash vector to all indexed MinHash vectors.
- Parameters
-
vec (
VectorUint) – The query MinHash vector - Returns
-
The Jaccard distances
- Return type
-
Listoffloat
-
get_all_nearest_neighbors(self: tmap.LSHForest, k: int, kc: int=10) → tmap.VectorUint¶ -
Get the k-nearest neighbors of all indexed MinHash vectors.
- Parameters
-
k (
int) – The number of nearest neighbors to be retrieved - Keyword Arguments
-
kc (
int) – The factor by whichkis multiplied for LSH forest retreival - Returns
-
VectorUintThe ids of all k-nearest neighbors
-
get_distance(self: tmap.LSHForest, arg0: tmap.VectorUint, arg1: tmap.VectorUint) → float¶ -
Calculate the Jaccard distance between two MinHash vectors.
- Parameters
-
-
vec_a (
VectorUint) – A MinHash vector -
vec_b (
VectorUint) – A MinHash vector
-
- Returns
-
floatThe Jaccard distance
-
get_distance_by_id(self: tmap.LSHForest, arg0: int, arg1: int) → float¶ -
Calculate the Jaccard distance between two indexed MinHash vectors.
- Parameters
-
-
a (
int) – The id of an indexed MinHash vector -
b (
int) – The id of an indexed MinHash vector
-
- Returns
-
floatThe Jaccard distance
-
get_hash(self: tmap.LSHForest, arg0: int) → tmap.VectorUint¶ -
Retrieve the MinHash vector of an indexed entry given its index. The index is defined by order of insertion.
- Parameters
-
a (
int) – The id of an indexed MinHash vector - Returns
-
VectorUintThe MinHash vector
-
get_knn_graph(self: tmap.LSHForest, from: tmap.VectorUint, to: tmap.VectorUint, weight: tmap.VectorFloat, k: int, kc: int=10) → None¶ -
Construct the k-nearest neighbor graph of the indexed MinHash vectors. It will be written to out parameters
from,to, andweightas an edge list.- Parameters
-
-
from (
VectorUint) – A vector to which the ids for the from vertices are written -
to (
VectorUint) – A vector to which the ids for the to vertices are written -
weight (
VectorFloat) – A vector to which the edge weights are written -
k (
int) – The number of nearest neighbors to be retrieved during the construction of the k-nearest neighbor graph
-
- Keyword Arguments
-
kc (
int) – The factor by whichkis multiplied for LSH forest retreival
-
get_weighted_distance(self: tmap.LSHForest, arg0: tmap.VectorUint, arg1: tmap.VectorUint) → float¶ -
Calculate the weighted Jaccard distance between two MinHash vectors.
- Parameters
-
-
vec_a (
VectorUint) – A weighted MinHash vector -
vec_b (
VectorUint) – A weighted MinHash vector
-
- Returns
-
floatThe Jaccard distance
-
get_weighted_distance_by_id(self: tmap.LSHForest, arg0: int, arg1: int) → float¶ -
Calculate the Jaccard distance between two indexed weighted MinHash vectors.
- Parameters
-
-
a (
int) – The id of an indexed weighted MinHash vector -
b (
int) – The id of an indexed weighted MinHash vector
-
- Returns
-
floatThe weighted Jaccard distance
-
index(self: tmap.LSHForest) → None¶ -
Index the LSH forest. This has to be run after each time new MinHashes were added.
-
is_clean(self: tmap.LSHForest) → bool¶ -
Returns a boolean indicating whether or not the LSH forest has been indexed after the last MinHash vector was added.
- Returns
-
Trueifindex()has been run since MinHash vectors have last been added usingadd()orbatch_add().Falseotherwise - Return type
-
bool
-
linear_scan(self: tmap.LSHForest, vec: tmap.VectorUint, indices: tmap.VectorUint, k: int=10) → List[Tuple[float, int]]¶ -
Query a subset of indexed MinHash vectors using linear scan.
- Parameters
-
-
vec (
VectorUint) – The query MinHash vector -
indices (
VectorUint) –
-
- Keyword Arguments
-
k (
int) – The number of nearest neighbors to be retrieved - Returns
-
The results of the query
- Return type
-
ListofTuple[float, int]
-
query(self: tmap.LSHForest, arg0: tmap.VectorUint, arg1: int) → tmap.VectorUint¶ -
Query the LSH forest for k-nearest neighbors.
- Parameters
-
-
vec (
VectorUint) – The query MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Returns
-
The results of the query
- Return type
-
VectorUint
-
query_by_id(self: tmap.LSHForest, arg0: int, arg1: int) → tmap.VectorUint¶ -
Query the LSH forest for k-nearest neighbors.
- Parameters
-
-
id (
int) – The id of an indexed MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Returns
-
The results of the query
- Return type
-
VectorUint
-
query_exclude(self: tmap.LSHForest, arg0: tmap.VectorUint, arg1: tmap.VectorUint, arg2: int) → tmap.VectorUint¶ -
Query the LSH forest for k-nearest neighbors.
- Parameters
-
-
vec (
VectorUint) – The query MinHash vector -
exclude (
ListofVectorUint) – -
k (
int) – The number of nearest neighbors to be retrieved
-
- Returns
-
The results of the query
- Return type
-
VectorUint
-
query_exclude_by_id(self: tmap.LSHForest, arg0: int, arg1: tmap.VectorUint, arg2: int) → tmap.VectorUint¶ -
Query the LSH forest for k-nearest neighbors.
- Parameters
-
-
id (
int) – The id of an indexed MinHash vector -
exclude (
ListofVectorUint) – -
k (
int) – The number of nearest neighbors to be retrieved
-
- Returns
-
The results of the query
- Return type
-
VectorUint
-
query_linear_scan(self: tmap.LSHForest, vec: tmap.VectorUint, k: int, kc: int=10) → List[Tuple[float, int]]¶ -
Query k-nearest neighbors with a LSH forest / linear scan combination.
k`*:obj:`kcnearest neighbors are searched for using LSH forest; from these, theknearest neighbors are retrieved using linear scan.- Parameters
-
-
vec (
VectorUint) – The query MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Keyword Arguments
-
kc (
int) – The factor by whichkis multiplied for LSH forest retreival - Returns
-
The results of the query
- Return type
-
ListofTuple[float, int]
-
query_linear_scan_by_id(self: tmap.LSHForest, id: int, k: int, kc: int=10) → List[Tuple[float, int]]¶ -
Query k-nearest neighbors with a LSH forest / linear scan combination.
k`*:obj:`kcnearest neighbors are searched for using LSH forest; from these, theknearest neighbors are retrieved using linear scan.- Parameters
-
-
id (
int) – The id of an indexed MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Keyword Arguments
-
kc (
int) – The factor by whichkis multiplied for LSH forest retreival - Returns
-
The results of the query
- Return type
-
ListofTuple[float, int]
-
query_linear_scan_exclude(self: tmap.LSHForest, vec: tmap.VectorUint, k: int, exclude: tmap.VectorUint, kc: int=10) → List[Tuple[float, int]]¶ -
Query k-nearest neighbors with a LSH forest / linear scan combination.
k`*:obj:`kcnearest neighbors are searched for using LSH forest; from these, theknearest neighbors are retrieved using linear scan.- Parameters
-
-
vec (
VectorUint) – The query MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Keyword Arguments
-
-
exclude (
ListofVectorUint) – -
kc (
int) – The factor by whichkis multiplied for LSH forest retreival
-
- Returns
-
The results of the query
- Return type
-
ListofTuple[float, int]
-
query_linear_scan_exclude_by_id(self: tmap.LSHForest, id: int, k: int, exclude: tmap.VectorUint, kc: int=10) → List[Tuple[float, int]]¶ -
Query k-nearest neighbors with a LSH forest / linear scan combination.
k`*:obj:`kcnearest neighbors are searched for using LSH forest; from these, theknearest neighbors are retrieved using linear scan.- Parameters
-
-
id (
int) – The id of an indexed MinHash vector -
k (
int) – The number of nearest neighbors to be retrieved
-
- Keyword Arguments
-
-
exclude (
ListofVectorUint) – -
kc (
int) – The factor by whichkis multiplied for LSH forest retreival
-
- Returns
-
The results of the query
- Return type
-
ListofTuple[float, int]
-
restore(self: tmap.LSHForest, arg0: str) → None¶ -
Deserializes a previously serialized (using
store()) state into this instance ofLSHForestand recreates the index.- Parameters
-
path (
str) – The path to the file which is deserialized
-
size(self: tmap.LSHForest) → int¶ -
Returns the number of MinHash vectors in this LSHForest instance.
- Returns
-
The number of MinHash vectors
- Return type
-
int
Layout
- layout_from_lsh_forest(lsh_forest: tmap::LSHForest, config: tmap.LayoutConfiguration=tmap.LayoutConfiguration(), create_mst: bool=True, clear_lsh_forest: bool=False, weighted: bool=False) → Tuple[tmap.VectorFloat, tmap.VectorFloat, tmap.VectorUint, tmap.VectorUint, tmap.GraphProperties]
-
Create minimum spanning tree or k-nearest neighbor graph coordinates and topology from an
LSHForestinstance.- Parameters
- Keyword Arguments
-
-
config (
LayoutConfiguration, optional) – AnLayoutConfigurationinstance -
create_mst (
bool): Whether to create a minimum spanning tree or to return coordinates and topology for the k-nearest neighbor graph clear_lsh_forest (bool) – Whether to runclear()on theLSHForestinstance after k-nearest neighbor graph and MST creation and before layout
-
config (
- Returns
-
The x and y coordinates of the vertices, the ids of the vertices spanning the edges, and information on the graph
- Return type
-
Tuple[VectorFloat, VectorFloat, VectorUint, VectorUint, GraphProperties]
- layout_from_edge_list(vertex_count: int, edges: List[Tuple[int, int, float]], config: tmap.LayoutConfiguration=tmap.LayoutConfiguration(), create_mst: bool=True) → Tuple[tmap.VectorFloat, tmap.VectorFloat, tmap.VectorUint, tmap.VectorUint, tmap.GraphProperties]
-
Create minimum spanning tree or k-nearest neighbor graph coordinates and topology from an edge list.
- Parameters
-
vertex_count (
int): The number of vertices in the edge list edges (ListofTuple[int, int, float]) – An edge list defining a graph - Keyword Arguments:
-
-
config (
LayoutConfiguration, optional) – AnLayoutConfigurationinstance -
create_mst (
bool) – Whether to create a minimum spanning tree or to return coordinates and topology for the k-nearest neighbor graph
-
config (
- Returns
-
The x and y coordinates of the vertices, the ids of the vertices spanning the edges, and information on the graph
- Return type
-
Tuple[VectorFloat, VectorFloat, VectorUint, VectorUint, GraphProperties]
-
tmap.mst_from_lsh_forest(lsh_forest: tmap::LSHForest, k: int, kc: int=10, weighted: bool=False) → Tuple[tmap.VectorUint, tmap.VectorUint]¶ -
Create minimum spanning tree topology from an
LSHForestinstance.- Parameters
- Keyword Arguments
-
-
kc (int) – The scalar by which k is multiplied before querying the LSH forest. The results are then ordered decreasing based on linear-scan distances and the top k results returned
-
weighted (
bool) – Whether the MinHash vectors in theLSHForestinstance are weighted
-
- Returns
-
the topology of the minimum spanning tree of the data indexed in the LSH forest
- Return type
-
Tuple[VectorUint, VectorUint]
-
class
tmap.ScalingType(self: tmap.ScalingType, arg0: int) → None¶ -
The scaling types available in OGDF. The class is to be used as an enum.
Notes
The available values are
ScalingType.Absolute: Absolute factor, can be used to scale relative to level size change.ScalingType.RelativeToAvgLength: Scales by a factor relative to the average edge weights.ScalingType.RelativeToDesiredLength: Scales by a factor relative to the disired edge length.ScalingType.RelativeToDrawing: Scales by a factor relative to the drawing.
-
class
tmap.Placer(self: tmap.Placer, arg0: int) → None¶ -
The places available in OGDF. The class is to be used as an enum.
Notes
The available values are
Placer.Barycenter: Places a vertex at the barycenter of its neighbors’ position.Placer.Solar: Uses information of the merging phase of the solar merger. Places a new vertex on the direct line between two suns.Placer.Circle: Places the vertices in a circle around the barycenter and outside of the current drawingPlacer.Median: Places a vertex at the median position of the neighbor nodes for each coordinate axis.Placer.Random: Places a vertex at a random position within the smallest circle containing all vertices around the barycenter of the current drawing.Placer.Zero: Places a vertex at the same position as its representative in the previous level.
-
class
tmap.Merger(self: tmap.Merger, arg0: int) → None¶ -
The mergers available in OGDF. The class is to be used as an enum.
Notes
The available values are
Merger.EdgeCover: Based on the matching merger. Computes an edge cover such that each contained edge is incident to at least one unmatched vertex. The cover edges are then used to merge their adjacent vertices.Merger.LocalBiconnected: Based on the edge cover merger. Avoids distortions by checking whether biconnectivity will be lost in the local neighborhood around the potential merging position.Merger.Solar: Vertices are partitioned into solar systems, consisting of sun, planets and moons. The systems are then merged into the sun vertices.Merger.IndependentSet: Uses a maximal independent set filtration. See GRIP for details.
-
class
tmap.LayoutConfiguration(self: tmap.LayoutConfiguration) → None¶ -
A container for configuration options for
layout_from_lsh_forest()andlayout_from_edge_list().-
int k -
The number of nearest neighbors used to create the k-nearest neighbor graph.
- Type
-
int
-
int kc -
The scalar by which k is multiplied before querying the LSH forest. The results are then ordered decreasing based on linear-scan distances and the top k results returned.
- Type
-
int
-
int fme_iterations -
Maximum number of iterations of the fast multipole embedder.
- Type
-
int
-
bool fme_randomize -
Whether or not to randomize the layout at the start.
- Type
-
bool
-
int fme_threads -
The number of threads for the fast multipole embedder.
- Type
-
int
-
int fme_precision -
The number of coefficients of the multipole expansion.
- Type
-
int
-
int sl_repeats -
The number of repeats of the scaling layout algorithm.
- Type
-
int
-
int sl_extra_scaling_steps -
Sets the number of repeats of the scaling.
- Type
-
int
-
double sl_scaling_min -
The minimum scaling factor.
- Type
-
float
-
double sl_scaling_max -
The maximum scaling factor.
- Type
-
float
-
ScalingType sl_scaling_type -
Defines the (relative) scale of the graph.
- Type
-
int mmm_repeats -
Number of repeats of the per-level layout algorithm.
- Type
-
int
-
Placer placer -
The method by which the initial positons of the vertices at eachlevel are defined.
- Type
-
Merger merger -
The vertex merging strategy applied during the coarsening phaseof the multilevel algorithm.
- Type
-
double merger_factor -
The ratio of the sizes between two levels up to which the mergingis run. Does not apply to all merging strategies.
- Type
-
float
-
int merger_adjustment -
The edge length adjustment of the merging algorithm. Does notapply to all merging strategies.
- Type
-
int
-
float node_size -
The size of the nodes, which affects the magnitude of their repellingforce. Decreasing this value generally resolves overlaps in a verycrowded tree.
- Type
-
float
Constructor for the class
LayoutConfiguration.-
__init__(self: tmap.LayoutConfiguration) → None¶ -
Constructor for the class
LayoutConfiguration.
-
-
class
tmap.GraphProperties(self: tmap.GraphProperties) → None¶ -
Contains properties of the minimum spanning tree (or forest) generated by
layout_from_lsh_forest()andlayout_from_edge_list().-
mst_weight¶ -
The total weight of the minimum spanning tree.
- Type
-
float
-
n_connected_components¶ -
The number of connected components in the minimum spanning forest.
- Type
-
int
-
n_isolated_vertices¶ -
- Type
-
int
-
degrees¶ -
The degrees of all vertices in the minimum spanning tree (or forest).
- Type
-
VectorUint
-
adjacency_list¶ -
The adjaceny lists for all vertices in the minimum spanning tree (or forest).
- Type
-
ListofVectorUint
Constructor for the class
GraphProperties.-
__init__(self: tmap.GraphProperties) → None¶ -
Constructor for the class
GraphProperties.
-